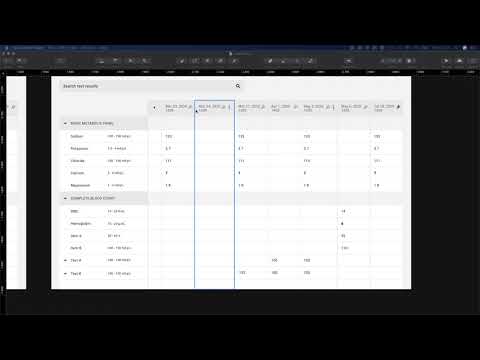
We saw a proposed design for a results view concept for micro frontends on today’s Squad Showcase:
While there are some nice ideas here (e.g., pinning a column while browsing tabular data), there are several issues as well (using only bold for abnormals, focused on numeric results, extra clicks & popups for graphs & detail, etc.). While I dumped some feedback into the #mf-test-results-viewer channel on Slack, where it can fall into the void of never-to-be-seen-again Slack chat history, I thought it was worth some more design discussion, so I’m bringing here to our community forum (aka mailing list).
At Regenstrief, I led an effort to create a tool called “Chart Search” that took an approach to results review combining two goals:
- Let me “google” the patient’s chart
- When a user selects a result, show them everything they might want/need to know about that result.
We used Lucene to index the patients chart, which provided instant results, counts for matches by result type, and support for synonyms & context-highlighting within text. Our chart search windows comprised (1) search & filtering, (2) a list of results, and (3) a detail pane to show detail for the currently selected result. I’ll share some screen shots…
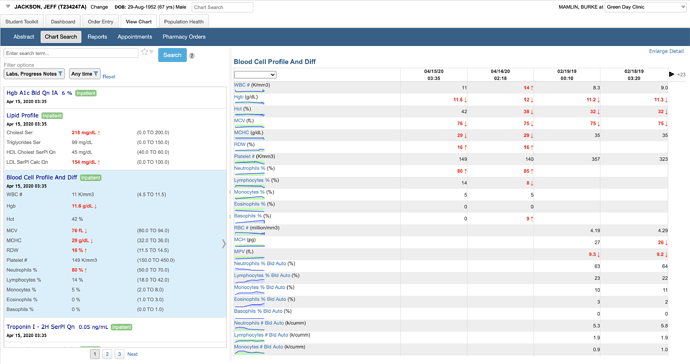
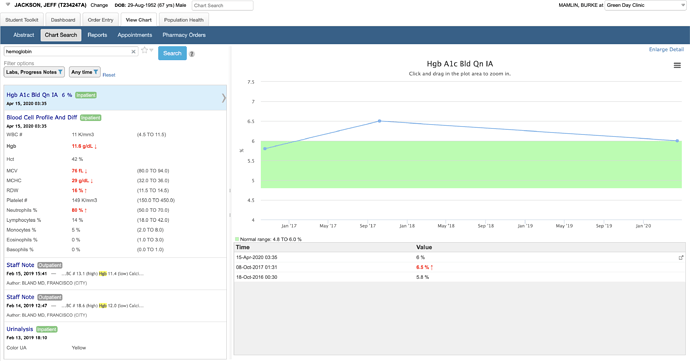
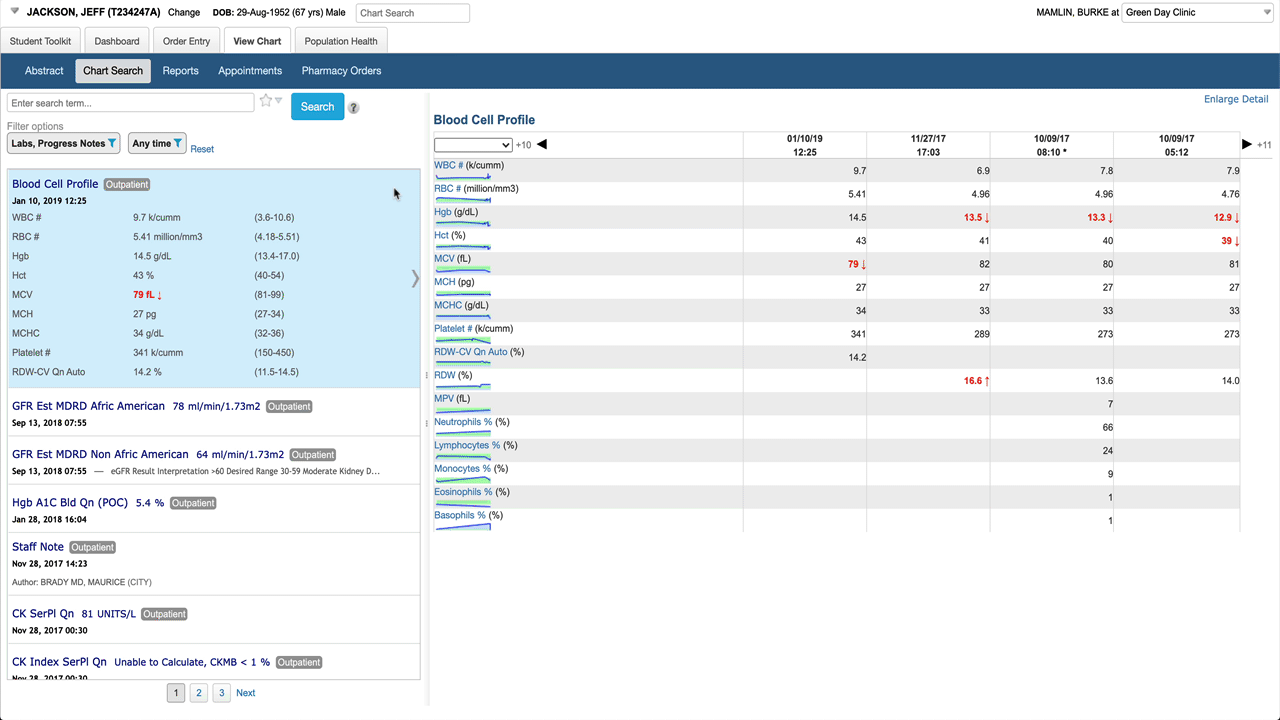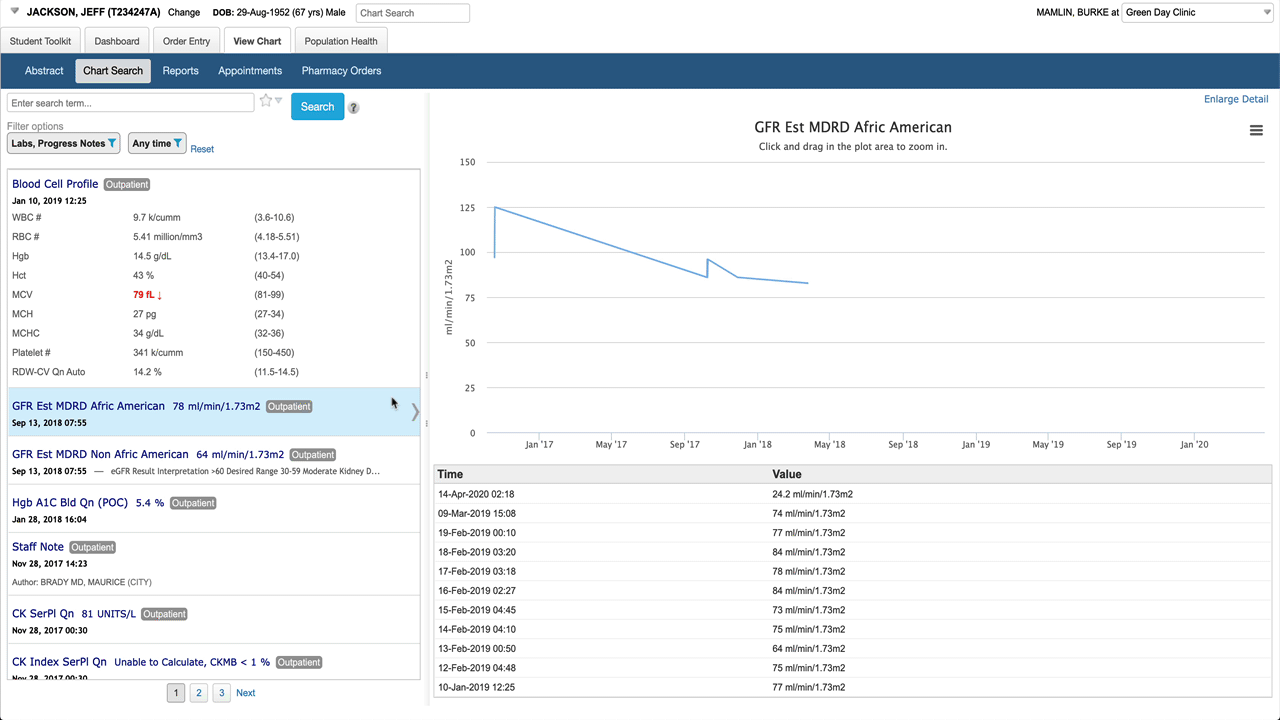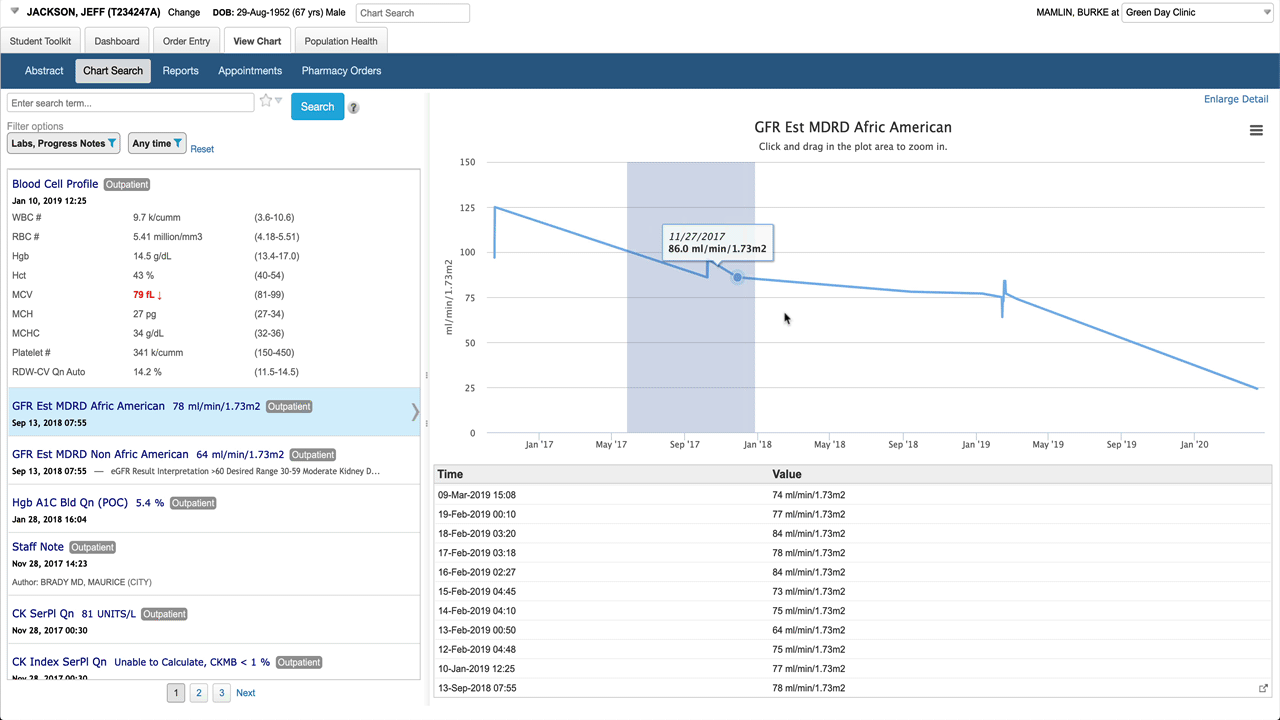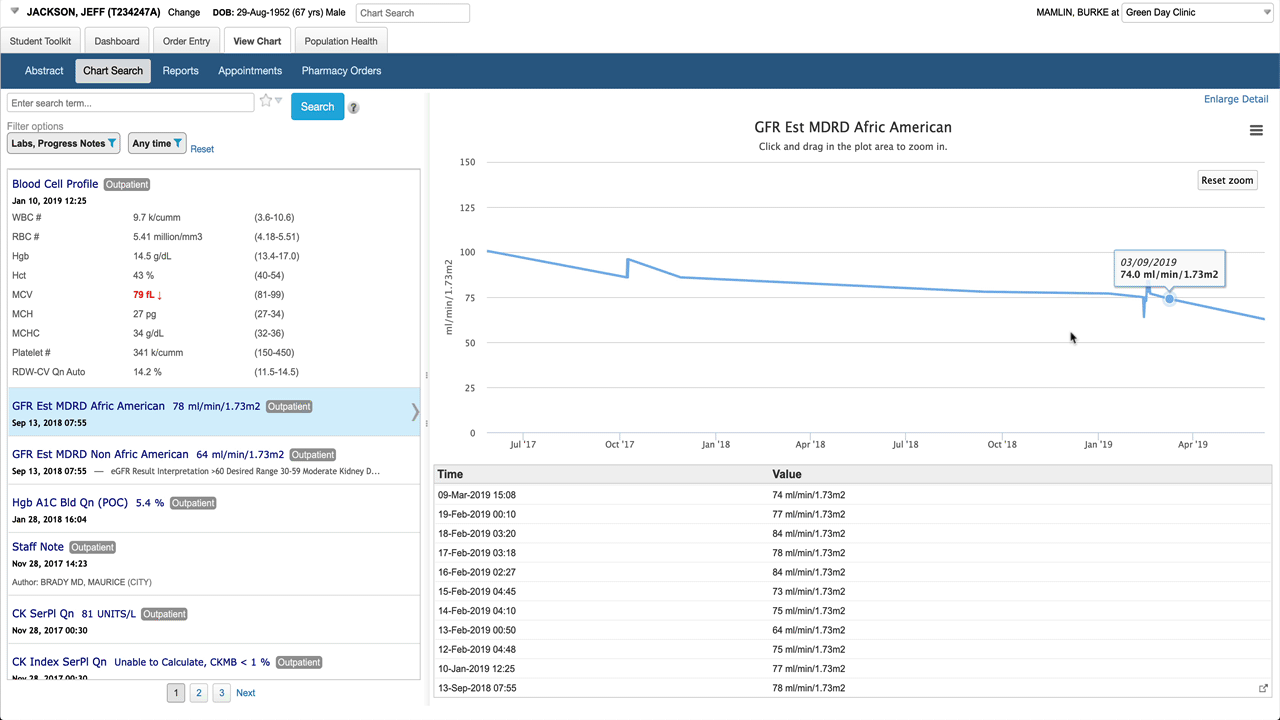
Here’s the basic layout. When a battery (lab result with multiple components) is selected in the result list, the detail pane shows a table by default. Note abnormals use more than just bold/color to be more accessible, spark lines give a preview of a graph for each result. Clicking on any result in the table shows detail about that specific component.
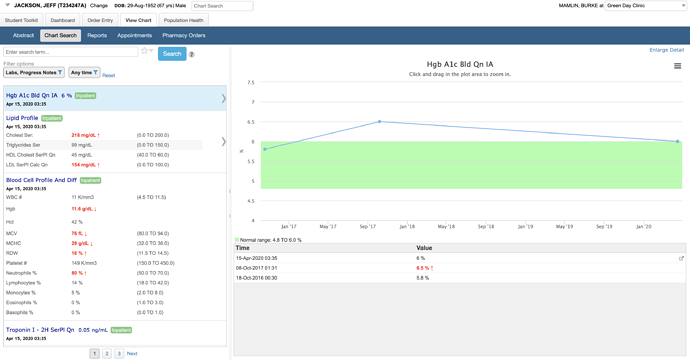
Selecting an individual numeric result shows a graph and a list of values in the detail pane.
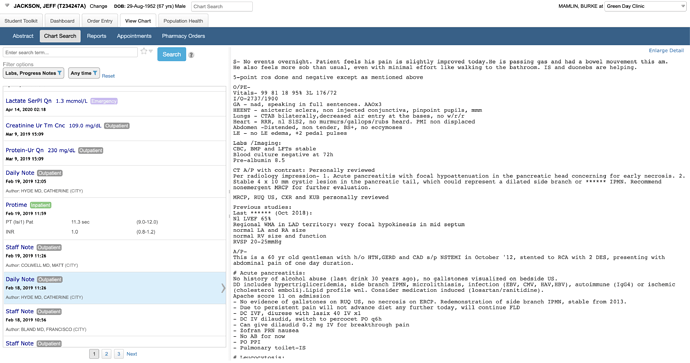
Results are not limited to labs or just numeric data. You are effectively googling the patient’s chart.
This example shows a search for “hemoglobin” with support for synonyms and context highlighting in text:
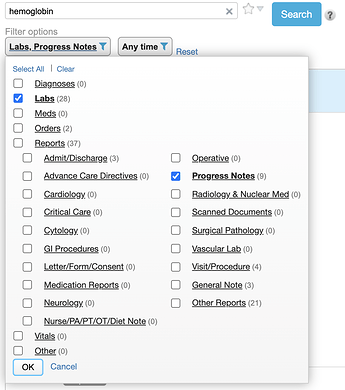
You can quickly filter and know where the data are – e.g., in this example I know there are 28 labs, 3 admit/discharge notes, and no cardiology reports mentioning hemoglobin before I make any filter selections. I can randomly select multiple with checkboxes or instantly narrow to one by clicking on its name. I can also save my default set of filters. In a different version of Chart Search, we let you save specific filter settings as quick links.
@k.joseph did a valiant effort of introducing a Chart Search module for the Reference Application; however, we never got it refined enough to be useful for implementation for wide adoption.