Globally, significant progress has been made in enrolling more HIV+ persons in lifesaving antiretroviral therapy (ART). According to UNAIDS figures, 27.5 million people were receiving HIV treatment at the end of 2020, up from 7.8 million in 2010, including more than one million in Nigeria. While more patients have been enrolled in treatment, long-term retention remains one of the most challenging, posing a significant risk to the 95-95-95 goals of ending the HIV epidemic by 2030.
The highest concentration of interruption in treatment (IIT) or loss to follow-up (LTFU) and other pain points have been seen in the first few months of HIV treatment. To achieve HIV epidemic control, Public Health Information, Surveillance Solutions, and Systems (PHIS3) is developing a predictive model for IIT/LTFU among clients on HIV treatment in Nigeria using historical data available on the NigeriaMRS (OpenMRS). The machine learning predictive model will help HIV program stakeholders in identifying clients with a high likelihood of IIT in order to guide preemptive efforts to avert the expected outcome. Python is the programming language used to create the model, which is subsequently displayed in charts and tables on the NigeriaMRS patient dashboard.
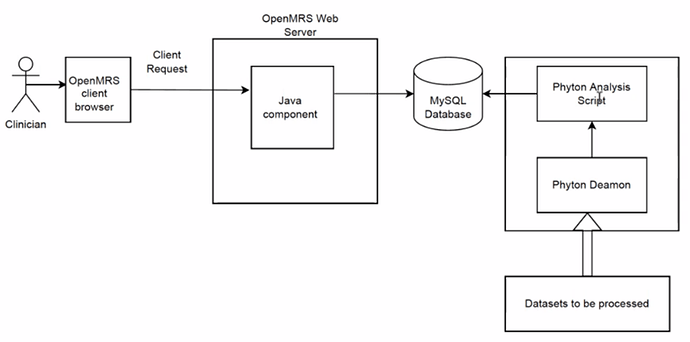
The model building is completed and integration of the NigeriaMRS is ongoing. See the schematic diagram below.
Figure 1: Shows a schematic diagram of the project
…
…
Best regards,
Gibril Gomez