Currently we are using EmrConceptSearchController from EMR API for diagnosis search which internally calls EmrConceptService.conceptSearch(…). This the same API used for diagnosis search in visit note in OpenMRS ref app. However the difference is in the controller. In EmrConceptSearchController we are passing the concept sources as null and hence it does not search by reference terms. In reference app we pass “ICD-10-WHO” as source. Should we modify the controller or have a separate API which will call the conceptSearch(…) with sources in parameters? Also should the source always be “ICD-10-WHO” only?
@shruthipitta I would expect that “which reference source to search for diagnoses” is a one-off server-side configuration, and in fact I think we should use what the Reference Application already uses, i.e. org.openmrs.module.emrapi.EmrApiProperties#getConceptSourcesForDiagnosisSearch().
At present I see that it’s hardcoded to “ICD-10-WHO” but we should change the emrapi module to make it configurable (by a GlobalProperty or by Metadata Mapping), still defaulting to “ICD-10-WHO” for backwards-compatibility.
What do you think?
Limiting that search to one configurable source sounds like a good way to go.
@shruthipitta I think you should go ahead an mimic the Ref App’s behaviour. Then we should open an EMR API ticket to enhance this so that the source becomes configurable via a GP or by Metadata Mapping as @darius proposed.
Just created this:
- EA-130: 'Concept sources for diagnosis search to be configurable via GP or Metadata Mapping’.
The controller is also in EMR API EmrConceptSearchController. Should I raise a separate ticket and fix controller as part of that? Is it safe to modify the controller, I mean other implementation could be using it?
@shruthipitta it would be useful if you could provide a link to the concern line(s) on GitHub. You can use the permalink feature of GitHub for this. Example:
Yes you should assume that this controller is used. So backward compatibility should be of concern. Are there currently sufficient unit tests behind this controller?
Can you point me to what you envision to change?
The only behavior you’re adding is that searching will also search by reference terms (when configured)?
Personally I think it’s fine to just add this behavior to the controller without being concerned about backwards-compatibility. I.e. nobody would have been searching for a concept called “E11.9” by name before.
I am not sure I understand the usages pattern here. Are we saying that the user just types ‘Cholera’ or ‘A00’ and we show the matching diagnoses? How are we identifying whether to search on simple name or Code?
If its just by default, consider the performance aspect as well of doing this.
- Bahmni expects Diagnosis to be organized as ‘set of sets’. So, essentially, you have to search within the sets defined.
- If you are doing a like search against name and code, its got to be a union SQL. I would expect that to be really really slow.
- Consider this, Bangladesh has more than 10000 ICD-10 codes, and doing this is going to slow things down considerably. (btw, we just created synonyms prefixed with the ICD 10 code to solve the search problem)
For reference, this is [BAH-310] - Bahmni - JIRA (though there’s nothing useful written there to give context).
What I understand we’re doing is replicating the same functionality that the Reference Application already has in its own diagnosis widget, i.e. that the user may type in either the name or the code (typically ICD10 but this would be configurable).
In the refapp it looks like this for your example:
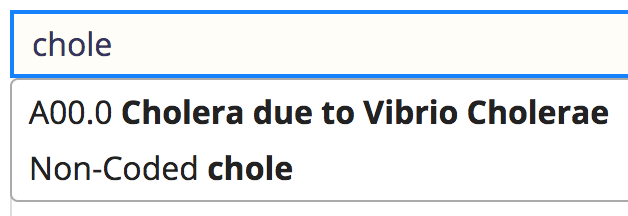
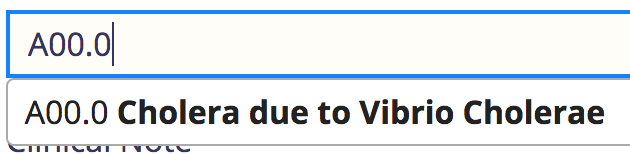
Note that it only does exact match on the code, or a like search by name.
I expect that @shruthipitta is not going to write any new back-end code for this but just use what’s in the emrapi module for the refapp. And I don’t think that PIH has ever complained about this code’s performance in their Mirebalais implementation, which has many diagnoses. (@ball, @ddesimone, @mogoodrich, does the diagnosis search widget in the Mirebalais visit note perform well?)
But yes, we should still test the performance of this against some realistic Bahmni dataset to see if we need to switch this over to search via lucene.
Been skimming this thread today and been meaning to follow up in greater details… but, as a quick answer, I believe we have thousands of diagnoses and as far as I know performance hasn’t been an issue… I don’t remember exactly how it works, but can hopefully have some time to glance at the code tomorrow.
Take care, Mark
As @darius mentioned, I will just use the same service API and instead of passing the sources as null I would pass EmrApiProperties#getConceptSourcesForDiagnosisSearch() as sources. [This] (https://github.com/openmrs/openmrs-module-emrapi/blob/42424342b36fdedd1febb15862eb79e13a1c26a9/omod/src/main/java/org/openmrs/module/emrapi/web/controller/EmrConceptSearchController.java#L49) is where the change would be.
@angshuonline search by name is a separate query which would run as previously. There is an extra query for matching the code with in the diagnosis set and this does is an EXACT match (does NOT have Like).
I would strongly advise against tying codes to the descriptions as these change over time. I favor being able to search based on reference code map. I would also point out that there indeed is a one-to-many relationship between disease and concept(s). It is not just enough to stem the ICD code or use the SNOMED hierarchy. There are a significant number of use cases where the roll-up of codes is not intuitive. Therefore there should also be a method of referencing a Set or Group which expands the interested concept into all the individual concepts necessary.
@akanter I did not completely understand . Can you please elaborate/ rephrase?
@angshuonline The extra query which I mentioned in my previous comment will run only if we pass any sources. This is determined by EmrApiProperties#getConceptSourcesForDiagnosisSearch(). Once EA-130: 'Concept sources for diagnosis search to be configurable via GP or Metadata Mapping’ is fixed an implementation can decide if they want to search by source/code or not. If they have a performance issue they can make the source as null so that the second query to fetch concepts by code will not run (like the current behaviour).
I am not sure about passing ‘sources’. How will we determine the code system? I am sure not defined by the users. If looked up from a global property, note there might be multiple sources defined. For example, Amputation of Lower Limb in ICD 9 is 84.1, the same is mapped in SNOMED CT to 397117006. So searching by 84.1 and 397117006 should result in the same. If we search against multiple sources, because a concept can have mappings to multiple concepts the queries are going to be extremely difficult to optimize. (and doing this using an ORM will not help). I would have preferred doing a lucene search, but lucene search does not categorize the diagnosis (set of sets defined)
First, the mappings between a concept and a code system are not equivalent. For example ICD is not the same conceptually as SNOMED, so just because a given concept is mapped to an ICD code and to a SNOMED CT code, that searching for that ICD code will return all the same concepts as searching for the SNOMED code. Each reference map has its own strengths and weaknesses and must be considered separately. Returning a concept based on its reference map code would need to know what the relationship type of the map is. For example, there really should only be ONE concept mapped SAME-AS a particular code. However, there will be generally MANY concepts mapped NARROWER-THAN to a code.
What I was describing above about code expansion can occur either because there are multiple concepts mapped to the exact same code, OR because you want to capture all of the descendant (children, grandchildren, etc.) of the code. So if you want all patients with Diabetes, you can search for all concepts mapped to ICD-10 E11.*. A generic patient with diabetes mellitus will be mapped to E11.9 but there will be no concepts mapped to just E11. There will be multiple different concepts mapped to E11.9 and there will be multiple concepts mapped to E11.0, E11.1, … etc. For SNOMED, the codes are not stemmed (so you can’t just truncate the code and look for groups of concepts). You have to use the SNOMED_RELATIONSHIPS to know which SNOMED concepts are more specific forms of Diabetes Mellitus. However, even that doesn’t always work since there are lots of concepts related to Diabetes in SNOMED that do NOT fall under the Diabetes Mellitus concept (they fall under a separate branch of SNOMED where complications of diabetes live).
@akanter the current behavior of this code (which is already in the reference application):
- We search by name in the standard OpenMRS way (so we can skip this)
- Searching by term will only return exact matches:
- “E14” matches nothing (since there are no concepts mapped to this exact term)
- “E14.*” matches nothing (since we don’t support this syntax)
- “E14.9” matches Diabetes Mellitus (this concept)
I believe matching like this satisfies your comments, right Andy?
But there’s one thing I see as a problem: currently our code doesn’t limit based on the map type. Usually this doesn’t matter because the CIEL dictionary’s diagnoses only have SAME-AS and NARROWER-THAN mappings. But we should be explicit about this, right? (And since I recall that these are standardized in OpenMRS we can hardcode against those exact strings, right?)
The answer in brief is YES. Depending on the search use case.