Thanks @burke for the response. Am following this up with @zacbutko on the pull request he raised to consume this data: [O3-1060] Data Timeline FilterSet by ZacButko · Pull Request #557 · openmrs/openmrs-esm-patient-chart · GitHub
Via REST, Obs group members are returned as the groupMembers property of the grouping Obs, so, e.g., to pick on one structure CIEL:1421 (Immunization history and how we implement Immunizations in the FHIR2 module) would result in something like:
{
concept: "1421AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA",
// other attributes
groupMembers: [
{
// CIEL:984 = Immunization given
concept: "984AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA",
// CIEL:5864 = Yellow fever
valueConcept: "5864AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA",
// other attributes
},
{
// CIEL:1420 Immunization sequence number
concept: 1420AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA,
valueNumeric: 1,
// other attributes
},
// other obs in this obs group
]
}
However, at least for the view discussed in this thread, we’d want to avoid pointing to obs groups at all if possible since we’re ultimately displaying the value obs and the handling of an obs group is likely to be more complex. E.g., the above example obs group really wants a custom view to translate the obs group into a single immunization record. This flattening of an obs group is obviously somewhat dependent on the structure of the obs group itself.
It seems possible to keep a relationship between a test data-point and an encounter. If this is passed through with the data the table could act as a hyperlink to navigate to the original encounter.
Does anyone know if this is desired behavior? From a front end perspective this is very quick to implement, given that the endpoint ties encounter data to each data point.
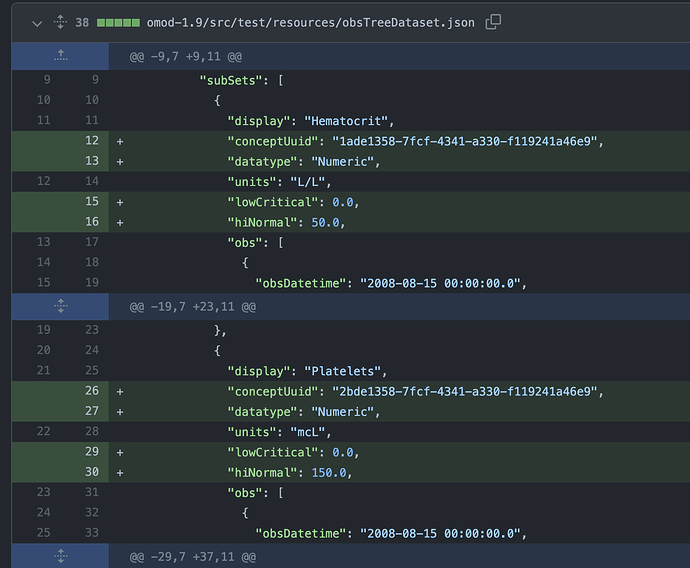
Regarding actual values of test ‘leaves’ I think this structure provides most of the functionality that the timeline needs. Here is an example for individual test type ‘Hematocrit’.
...
display: 'Complete Blood Count',
obs: [
{
display: 'Hematocrit',
conceptUuid: '1015AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA',
meta: {
datatype: 'Numeric',
hiAbsolute: 100,
hiCritical: null,
hiNormal: 51.9,
lowAbsolute: 0,
lowCritical: 21,
lowNormal: 32.3,
range: '32.3 – 51.9',
units: '%',
},
values: [
{
effectiveDateTime: '2020-05-07T00:00:00+00:00',
value: 42,
},
{
effectiveDateTime: '2020-05-07T00:02:00+00:00',
value: 43,
},
{
effectiveDateTime: '2020-05-08T00:03:00+00:00',
value: 45,
notes: 'Maybe there is some special note written here?',
},
],
},
{
display: 'Platelets',
...
},
...
@dkayiwa does this work for you?
@burke , @akanter , @ibacher are there some other data that needs to be displayed on Timeline table? Simplifying the data format to the above removes about 90% of information currently sent for the purpose of the timeline, but I believe these are actually the only information the timeline needs.
Yes it does. And in response to what you have just asked, i have made a followup commit at: RESTWS-882 add more concept numeric metadata · openmrs/openmrs-module-webservices.rest@26f6000 · GitHub
@zacbutko for the metadata, i only return those items that are not null. I did not return range because i did not see it in the data model. ![]()
Sorry if this is a silly question @dkayiwa - why are specific concepts showing up here in the PR? Are these just tests? We’re not hard-coding these, right? Sorry, I’m probably misunderstanding something…
e.g.
@grace the only silly question is the one which is not asked. ![]()
Yes you got it right that those are just tests, to help the reviewer see the output structure.
You can test the actual data at: https://qa-refapp.openmrs.org/openmrs/ws/rest/v1/obstree?patient=f83f4a1c-cc28-4a85-a8fb-bdf98130ec0e&concept=162552AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
You can replace the concept and patient parameters with your values. And you can optionally also supply the fromDate and toDate parameters.
Or use this interface: OpenMRS - Login with URI: /openmrs/ws/rest/v1/obstree?patient=f83f4a1c-cc28-4a85-a8fb-bdf98130ec0e&concept=162552AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
Thanks again @dkayiwa for being so accommodating of all these changes, and for getting the fixes out so quickly!
Range is an important field to give, if it exists for a test type. Below is some screenshot of how this is working right now on openmrs-spa.org. It seems that maybe this field does not always exist, but we should definitely include it if it does.
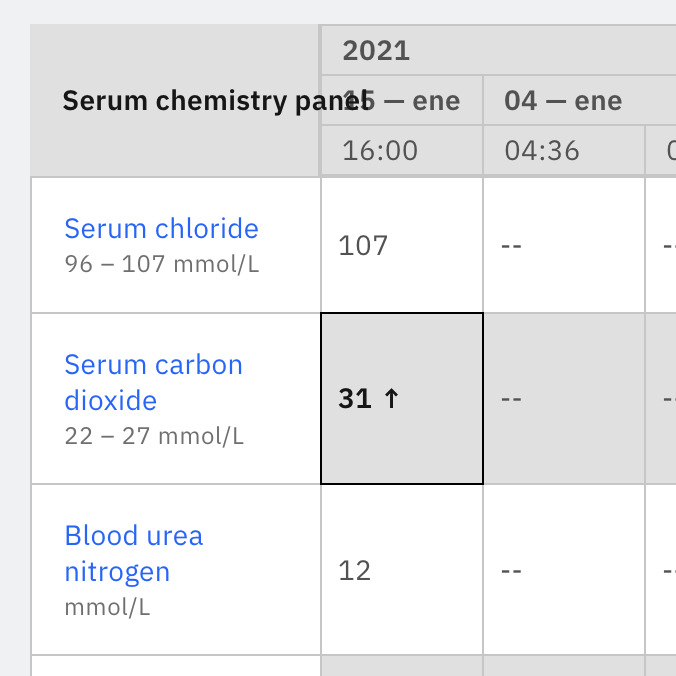
Could range be a combination of those fields? e.g lowNormal - hiNormal or lowAbsolute - hiAbsolute or lowCritical - hiCritical
Range is not a property provided by the backend. It’s metadata created in the frontend.
It’s lowNormal - hiNormal.
@dkayiwa @ibacher , ok yeah that makes sense. Sorry for assuming things about the data structure again! I can definitely do this on FE.
Tests with numeric results can have three thresholds set at the concept level:
- Absolute low and high - values outside of this range are considered invalid/impossible
- Critical low and high - values outside of this range need immediate attention
- Normal low and high - values inside this range are considered “normal”
In each case, a single value (low or high) indicates the other side is unbounded (for example, a normal high of 5 with no normal low would indicate normal is <5).
FYI - while OpenMRS has historically relied (and still relies) on these thresholds in the concept definition for determining the interpretation (e.g., normal, abnormal, critical), we eventually need to evolve to match the rest of the world where observations (the values) can declare their abnormality independent of the concept definition. In other words, the lab reporting a result declares whether the value is abnormal or critical rather than depending on us looking up ranges in our dictionary. This is because normal ranges can frequently vary depending on patient characteristics (gender, age, etc.). This may be a reason to migrate toward having the backend report abnormality (as obs interpretation in FHIR) rather than leaving it to the frontend.
One of the key elements here seems to be the ability to show the results in a flowsheet and potentially a graph. This requires that the values be comparable. One of the challenges with lab interoperability is that the standard codes (LOINC) do not ensure the same units of measure. Although they may differentiate between substance concentration (mmol) and mass concentration (mg), the results could be grams, milligrams, etc. There can also be multiple LOINCs which actually share comparable results. I would want to make sure that we have the correct sets which build up from the core result concepts through to the rows and the groups of rows. If this only needs to be robust enough to handle results from a single lab/concept dictionary then you can presume that the units of measure will be the same and only certain concepts will be used to capture, let’s say, a CBC. But if we need something more robust that will work across implementations, we might need to think about the dictionary design. I would want CIEL to support this fully. Let me know what we need to do.
@dkayiwa thank you for the rapid turnaround(s) on this feature. Outstanding work!