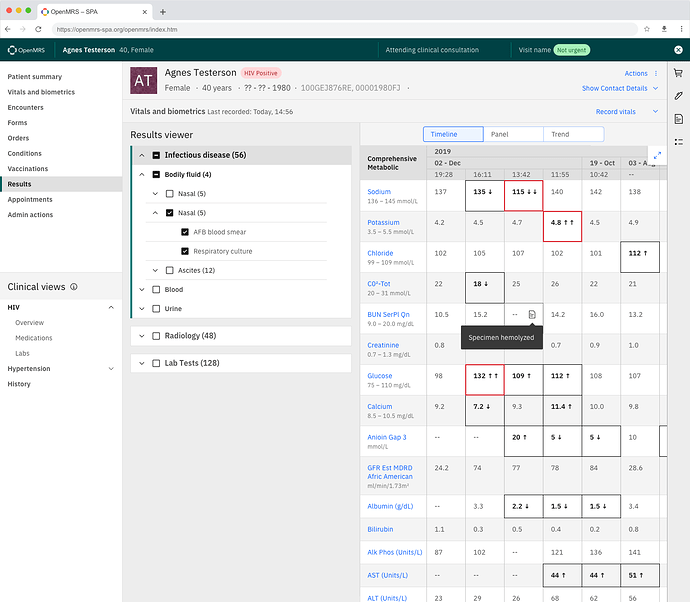
As we move forward with OpenMRS 3.0 features, the test results feature remains unfinished with regards to the designs.
The idea is to allow the user to have a filter tree to allow them to quickly move through categories of lab results. See https://app.zeplin.io/project/60d59321e8100b0324762e05/screen/60d5ea5bf120a6089c660a68
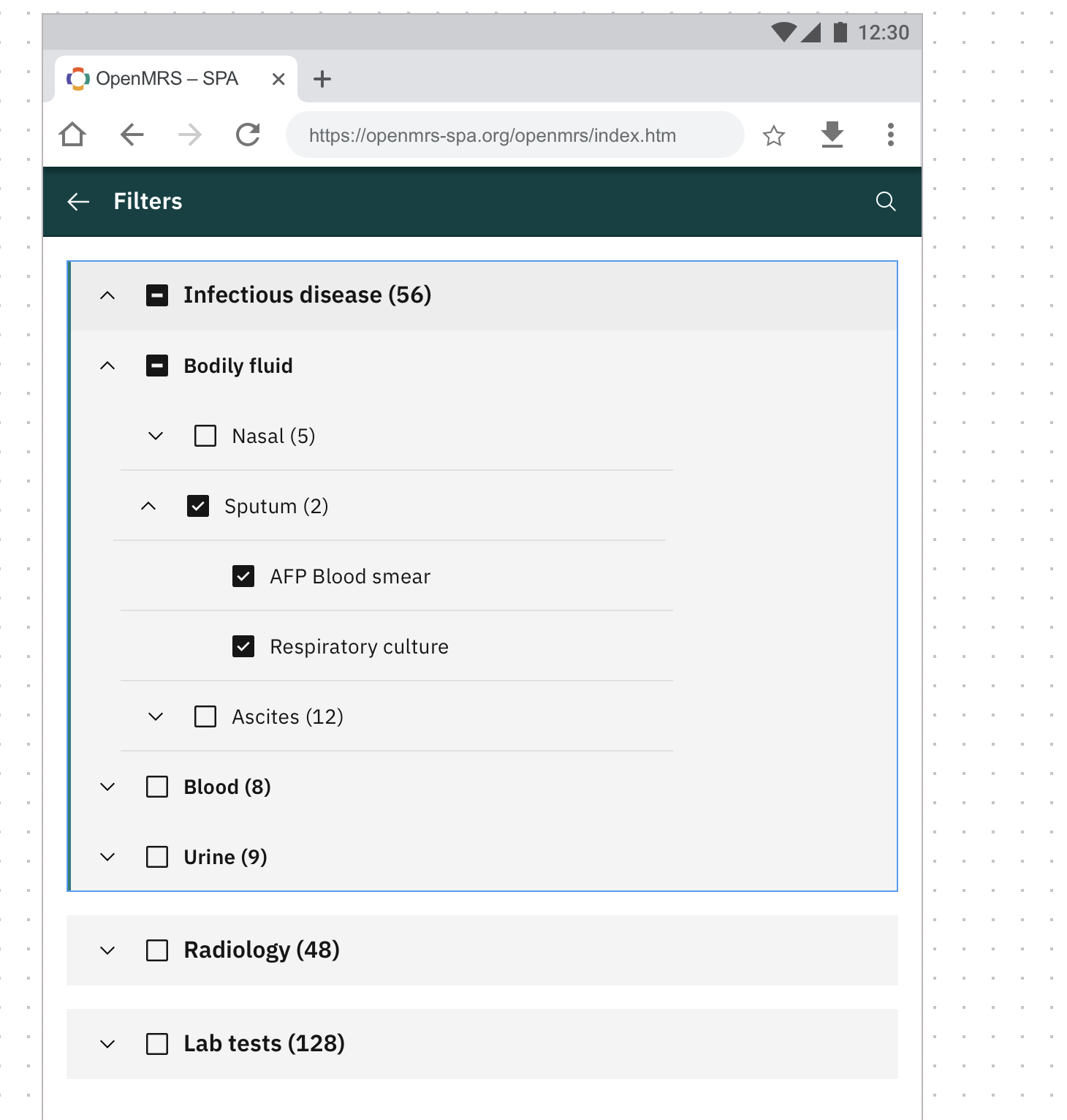
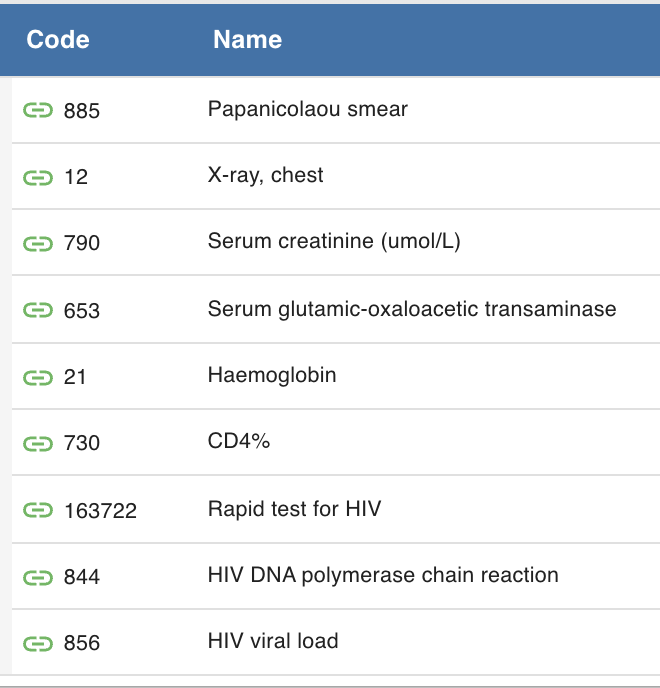
I would like to propose supporting this via concepts. A user would have the ability to create a concept set of concept sets based on their implementation needs. The leaf nodes of such. concept set would have to be of an allowable class (e.g. test result or test set).
To support, this I would like to propose the development of a new backend feature which takes a “root” concept, recursively unfolds the tree (could be done on initial load by OpenMRS if commonly used concepts are provided ahead of time) and on a rest request, if given a root concept, a patient id and a time window, will provide the “populated result” tree. The tree will be used to define the filters available in the UI such that only those Test with results are present.
Looking forward to hearing others’ thoughts.
JJ
@corneliouzbett @ibacher @Mekom @PIH @OHRI @burke @dkayiwa @aojwang (this is not meant to be exhaustive please make sure all are aware.